Farris Lab
-
Home Item
 Shannon Farris, Ph.D. , home
Shannon Farris, Ph.D. , homeAssistant Professor
Led by principal investigator Shannon Farris, Ph.D., the Farris lab studies the molecular and cellular mechanisms underlying learning. In particular, the lab is interested in understanding how neurons respond during learning and how those responses differ between genetically distinct cell types thought to be required for storing different parts of memory, such as who, what, when and where.
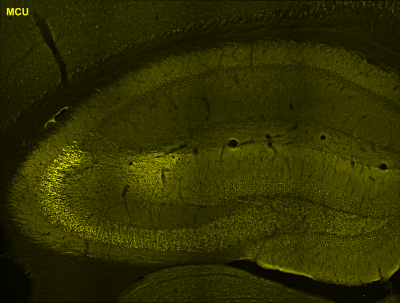
Neurons engaged during a learning experience often respond by altering the levels of transcription and translation of molecules needed for memory storage. One of the many challenges that neurons need to overcome is the distance between where molecules are made (in the nucleus) and where they are needed (at the synapse), which is often hundreds of microns away. One way neurons achieve this is by localizing RNA molecules to the synapse, so that they can be rapidly translated in response to local cues, such as changes in the activity of specific inputs. These newly synthesized proteins can then remodel the strength of synaptic connections in an input- and synapse-specific fashion. It’s widely believed that changes in the strength of specific synapses (both increases and decreases) are the molecular correlate of memory storage in the brain.
The Farris Lab uses a combination of mouse genetics to gain access to specific hippocampal cell-types, deep sequencing technologies to obtain a genome-wide view of transcription and translation, and single molecule imaging techniques to illuminate the processes regulating dendritic RNA in vivo. The lab aims to provide mechanistic insight on the behaviorally-induced synaptic modifications in CA2 that may be required for encoding social behavior.
Currently, the lab is focused on investigating dendritically localized RNAs in area CA2 of the hippocampus, a subregion that is resistant to long-term potentiation (synapse strengthening) and cell death, and is thought to be required for storing social memory, or “the who” aspect of memory. Given that a number of neurodevelopmental disorders are characterized by disruptions in social processing, investigating the post-transcriptional mechanisms critical to CA2 function and gaining a comprehensive molecular understanding of how social experiences are stored in the brain may lead to novel therapies relevant for individuals with social deficits.
Splicejam is a shiny-app resource that visualizes mouse hippocampal subregion- and compartment-specific RNA-seq data described in Farris et al. The purpose of this resource is to provide the community with a user-friendly interface to mine the data for isoform-specific differences across hippocampal subregions (CA1, CA2, CA3, DG) and subcellular compartments (Cell Body, Dendrites).
The RNA-seq data is visualized using sashimi plots that plot RNA-seq coverage data alongside splice junction-spanning sequence reads, using compressed intron genomic coordinates.
Click HERE to report bugs or issues with Splicejam.
- The methods were developed in support of S. Farris, J. M. Ward, K.E. Carstens, M. Samadi, Y. Wang and S. M. Dudek, 2019: Hippocampal subregions express distinct dendritic transcriptomes that reveal differences in mitochondrial function in CA2
- Sashimi plots were originally envisioned by MISO Katz, Y, Wang ET, Silterra J, Schwartz S, Wong B, Thorvaldsdóttir H, Robinson JT, Mesirov JP, Airoldi EM, Burge, CB.: Sashimi plots: Quantitative visualization of alternative isoform expression from RNA-seq data.
Splicejam is also available as an R package and is part of a suite of RNA-seq analysis R packages, collectively called “jampack” which is available through GitHub, see https://github.com/jmw86069/jampack
For vignettes on how to use Splicejam on other RNA-seq datasets go to: https://jmw86069.github.io/splicejam/
-
Bio Item
 Mayd Alsalman , bio
Mayd Alsalman , bioGraduate Student, Translational Biology, Medicine, and Health
-
Bio Item
 Mikel Cawley , bio
Mikel Cawley , bioGraduate Student, Translational Biology, Medicine, and Health
-
Bio Item
 Vernon Green , bio
Vernon Green , bioUndergraduate Student, Integrated Health Sciences Research
-
Bio Item
 Maanya Sappa , bio
Maanya Sappa , bioUndergraduate Student, Integrated Health Sciences Research
-
Bio Item
 Renesa Tarannum , bio
Renesa Tarannum , bioGraduate Student, Translational Biology, Medicine, and Health
-
Bio Item
 Lucy Turner , bio
Lucy Turner , bioResearch Assistant
-
Article Item
-
Article Item
-
Article Item
-
Article Item
-
Article Item
-
Article Item